字幕と単語
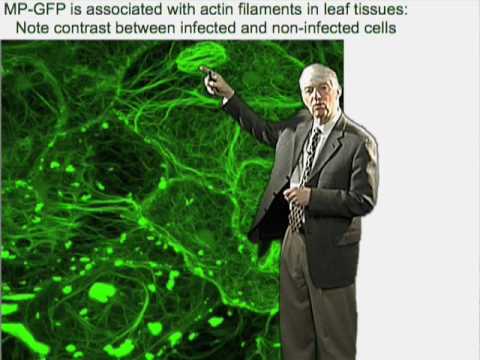
ロジャー・ビーチー(ダンフォースセンター) 第1部:植物ウイルス感染の生物学 (Roger Beachy (Danforth Center) Part 1: Biology of Plant Virus Infection)
00
江子揚 が 2021 年 01 月 14 日 に投稿保存
動画の中の単語
movement
US /ˈmuvmənt/
・
UK /ˈmu:vmənt/
- n. (c./u.)クラッシック音楽の楽章;同好、社会運動;社会的な運動;動き;(音楽の)楽章;運動;変化;機械;排便;(バレエの)動き;軍事行動
A2 初級
もっと見る エネルギーを使用
すべての単語を解除
発音・解説・フィルター機能を解除
